Near Telomere-to-telomere Japanese Reference Genome Sequence and Mitochondrial DNA Heteroplasmic Variants Panel Are Released on jMorp
The Japanese Reference Genome Sequence (JG3), the first Japanese near telomere-to-telomere (T2T) assembly, has been released. JG3 has improved accuracy compared to the previous version of the Japanese Reference Genome Sequence (JG2). In addition, a mitochondrial DNA heteroplasmic variants panel was also released, using whole genome analysis data from 8,380 Japanese people.
The JG3 and mitochondrial heteroplasmy panels are publicly available on the Japanese Multi Omics Reference Panel (jMorp), a database constructed by ToMMo.
Updated Details
Japanese Reference Genome Sequence; JG3
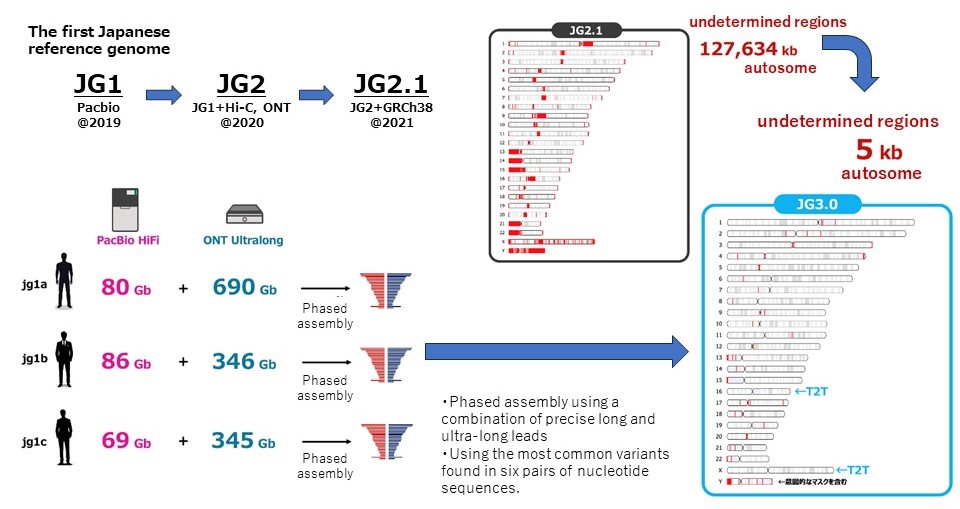
In 2022, the International Telomere to Telomere Consortium reported the full-length human genome sequence, proving that the whole human genome sequence can be determined. This prompted the ToMMo research team to work on the telomere-to-telomere genome assembly from blood samples taken from participants.
Through trial and error, a combination of two DNA sequencing technologies (Pacific Biosciences' HiFi reads and Oxford Nanopore Technologies' ultra-long DNA sequencing) has enabled the near telomere-to-telomere assembly. Based on this method, genome sequences were analyzed from three Japanese individuals. Six pairs of nucleotide sequences, representing almost all the nucleotide sequences of the original genome sequence, have been reconstructed from the DNA fragment data. Finally, a pair of near full-length genome sequences (Japanese Reference Genome Sequence; JG3) was constructed, adopting the major variants among the six sets of assemblies.
Approximately 5,100 base pairs remain undetermined, but this is significant progress compared to the previous 300 million base pairs of undetermined regions. There are also significant improvements in JG3. For example, the international reference genome sequence GRCh38 has incorrect duplicates of the KCNE1 gene, which causes heart failure when mutated, and the CBS gene, which causes homocystinuria when mutated, making analysis of this region difficult, but these errors have been corrected in JG3. It is expected to enable the analysis of previously impossible to analyze regions, and contribute to more accurate genome analysis.
Go to jMorp (JRGA (JG3): Japanese Reference Genome Sequence)
Dataset details (JG3.0.0)
Mitochondrial Heteroplasmy Panel
Genome sequencing data from 8,380 participants in the cohort study revealed for the first time aspects of mitochondrial heteroplasmic variants in the Japanese population. We found that mitochondrial heteroplasmy, although rare, is found in the general population. Although the haplogroups differ from European populations, the locations where heteroplasmy is observed are very similar. This suggests that there is a common mechanism for heteroplasmy to occur regardless of genetic origin.
Go to jMorp (Genome Variation (short-read WGS based mtDNA heteroplasmic variation panel))
Dataset details (8.3KJPN mtDNA variants panel)
How to use jMorp
Please refer to the link below.
https://jmorp.megabank.tohoku.ac.jp/help/tutorial
Related Link
Metabolome, transcriptome, metagenome, and genome data expansion -jMorp 2023 major update-(2023/09/04)
Constructing the First Version of the Japanese Reference Genome [Press Release](2021/01/29)

